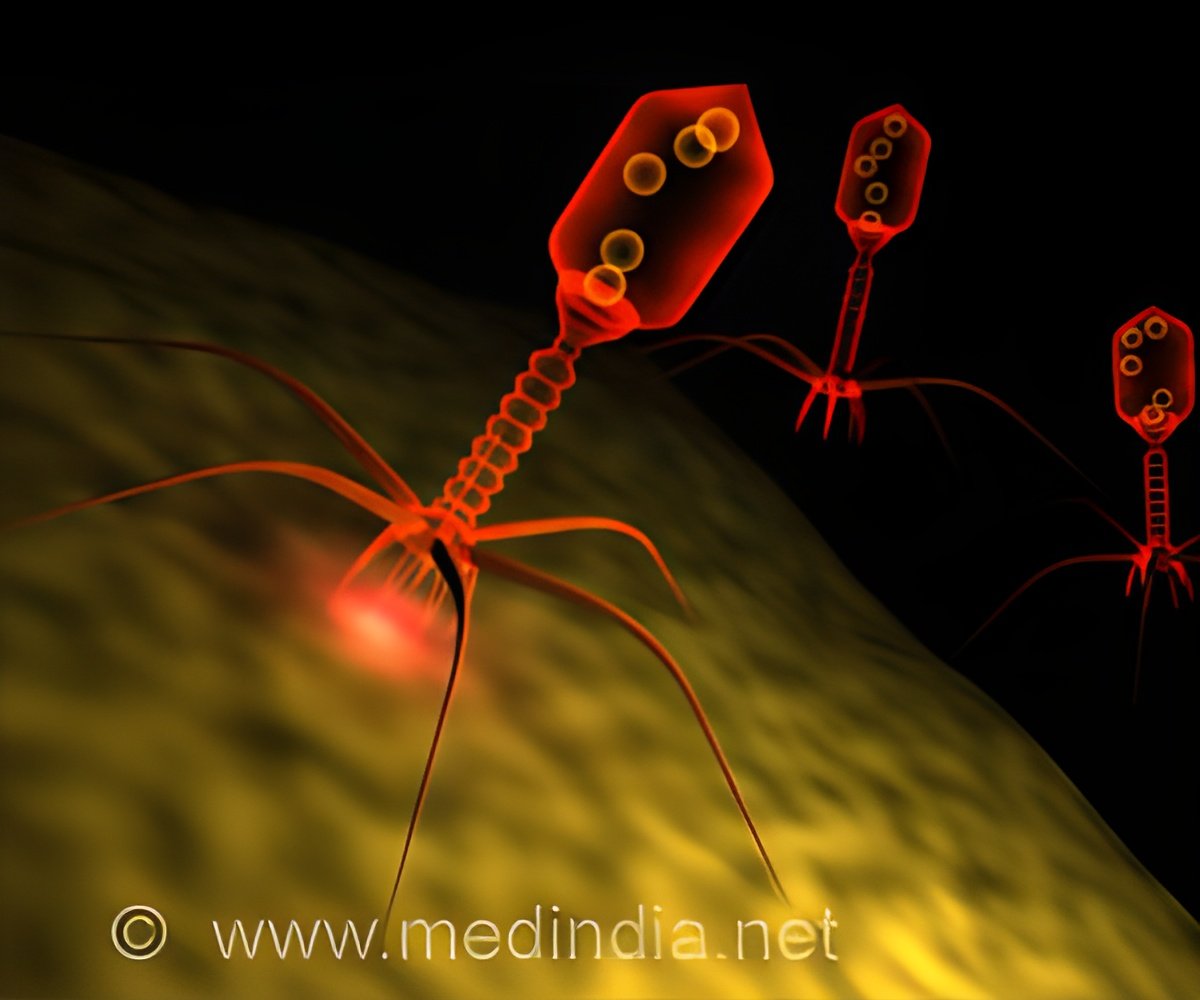
Bacteriophages, often known as phages, are viruses that engulf and kill bacteria. Although there are thousands of phages, it is difficult to use them as medicines to combat certain bacteria.
Scientists need to find approaches to turn phages into effective bacteria-killing devices to maximize phage therapy and make it scalable to treat human diseases.
This would also provide an alternative method of treating bacterial infections that are resistant to traditional antibiotics.
The medical and scientific sectors are looking for novel medications to combat infections as antibiotic resistance poses a more significant risk to human health.
A new method for utilizing the power of bacteriophages has brought researchers at Gladstone Institutes one step closer to their objective.
Gladstone scientists have developed an efficient method to alter phage genomes, enabling them to engineer new phages and target specific bacteria. This study was published in Nature Biotechnology( 1✔ ✔Trusted Source
Continuous multiplexed phage genome editing using recombitrons
Ultimately, finding a method and testing a large number of phage variants to identify the best ones will be crucial if phages are to be used to treat illnesses in patients who are resistant to many medications.
Several versions of the phage genome can be produced quickly and effectively by introducing various modifications to the genome using this novel approach.
The new approach relies on molecules called retrons, which originate from bacterial immune systems and act like DNA-production factories inside bacterial cells.
Shipman’s team has found ways to program retrons so they make copies of a desired DNA sequence. When phages infect a bacterial colony containing retrons, using the technique described in the team’s new study, the phages integrate the retron-produced DNA sequences into their genomes.
Advertisement
Bacteriophages in Antibiotic Resistance Treatments
Unlike antibiotics, which broadly kill many types of bacteria at once, phages are highly specific for individual strains of bacteria.
As rates of antibiotic-resistant bacterial infections rise—with an estimated 2.8 million such infections in the United States each year—researchers are increasingly looking at the potential of phage therapy as an alternative to combat these infections.
Already, phages have been successfully used in the clinic to treat a small number of patients with life-threatening antibiotic-resistant infections, but developing the therapies has been complex, time-consuming, and difficult to replicate at scale.
Doctors must screen collections of naturally occurring phages to test whether any could work against the specific bacteria isolated from an individual patient.
Shipman’s group wanted to find a way to modify phage genomes to create larger collections of phages that can be screened for therapeutic use, as well as to collect data on what makes some phages more effective or what makes them more or less specific to bacterial targets.
Advertisement
Recombitrons: Continuous Editing Tool for Phages
As the natural predators of bacteria, phages play an important role in shaping microbial communities, It’s important to have tools to modify their genomes to better study them.
It’s also important to engineer them so that microbial communities can be shaped to kill antibiotic-resistant bacteria.
To precisely engineer phage genomes, the scientists turned to retrons. In recent years, Shipman and his group pioneered the development and use of retrons to edit the DNA of human cells, yeast, and other organisms.
Shipman and his colleagues began by creating retrons that produce DNA sequences specifically designed to edit invading phages—a system the team dubbed “recombitrons.”
Advertisement
Efficient Multi-Gene Editing in Phages
Then, they put those retrons into colonies of bacteria. Finally, they let phages infect the bacterial colonies.
As the phages infected bacteria after bacteria, they continuously acquired and integrated the new DNA from the recombitrons, editing their genome as they went along.
The research team showed that the longer they let phages infect a recombitron-containing bacterial colony, the greater the number of phage genomes that were edited.
Moreover, the researchers could program different bacteria within the colony with different recombitrons, and the phages would acquire multiple edits as they infected the colony.
“As a phage is bouncing from bacterium to bacterium, it picks up different edits,” says Shipman.
Making multiple edits in phages is something that was previously incredibly hard to do; so much so that, most of the time, scientists simply didn’t do it.
Now, you throw some phages into these cultures, wait a while, and get your multiple-edited phages.
New Platform for Phage Screening
If scientists already knew exactly what edits they wanted to make to a given phage to optimize its therapeutic potential, the new platform would let them easily and effectively carry out those edits.
However, before researchers can predict the consequence of a genetic change, they first need to better understand what makes phages work and how variations to their genomes impact their effectiveness. The recombitron system helps make progress here, too.
If multiple recombitrons are put into a bacterial colony, and phages are allowed to infect the colony for only a short time, different phages will acquire different combinations of edits. Such diverse collections of phages could then be compared.
Scientists now have a way to edit multiple genes at once if they want to study how these genes interact or introduce modifications that could make the phage a more potent bacterial killer.
Shipman’s team is working on increasing the number of different recombitrons that can be put into a single bacterial colony—and then passed along to phages.
They expect that eventually, millions of combinations of edits could be introduced to phages to make huge screening libraries.
“We want to scale this high enough, with enough phage variants, that we can start to predict which phage variants will work against what bacterial infections,” says Shipman.
Reference:
- Continuous multiplexed phage genome editing using recombitrons – (https:www.nature.com/articles/s41587-024-02370-5)
Source-Eurekalert